At first to be able to rewrite the consensus pattern and use the motif5.pl to determine the N-glycosylation site, we change the motif to N[^^P][ST][^^P](for windows) and N[^P][ST][^P] (for Mac Computer).
a. Comparision of N-glycosylation sites obtained from motif5.pl and Prosite Scan of consensus pattern of given hypothetical protein.
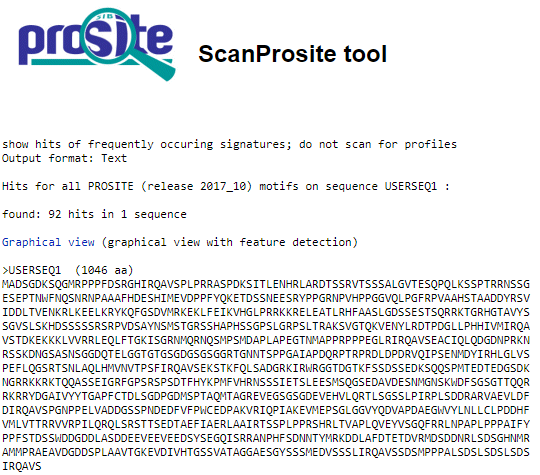

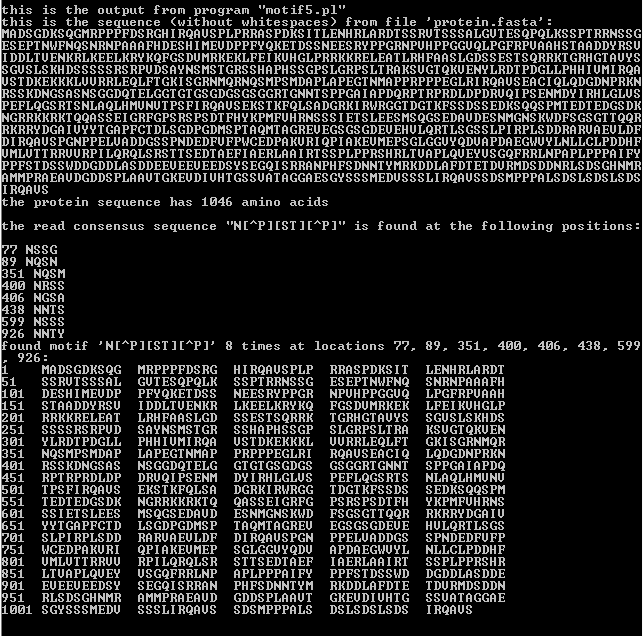
The result of N-glycosylation sites from Prosite Scan and output obtained by using motif5.pl is same. we found the motif 8 times at 77, 89, 351, 400, 406, 438, 599 and 926 positions.
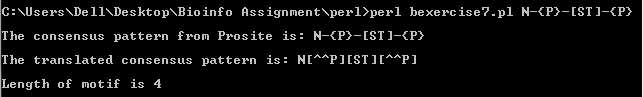
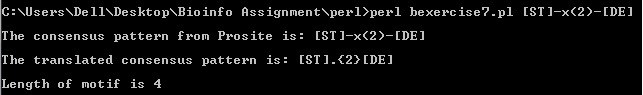
b. The perl program which is able to read the following consensus patterns
(i) N-{P}-[ST]-{P}
(ii)[ST]-X(2)-[DE]
(iii)X-G-[RK]-[RK]
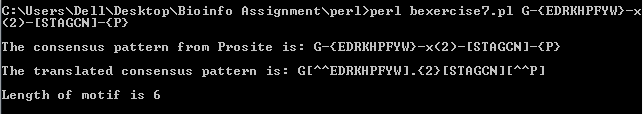
(IV)G-{EDRKHPFYW}-X(2)-[STAGON]-{P},
rewrite them into perl pattern and calculate the length of motifs is link to exercise 7b.
The output obtained from the perl program are as follows:

INFORMATION ON HOW TO USE THE PERL PROGRAM:
There is a perl program in the link to exercise7b. Click there, download the program and save it in ASCII format. After that go to command prompt, type dir, click enter then type cd (change directory), give a space and type the name of folder where the file is located and click enter, change directory until the final file location is reached. Then type final file name, give a space and type bexercise7.pl and type the given consensus pattern and enter, it will display the output.